Viper is transforming research at the University of Hull , listed below are examples of how researchers are using Viper to improve their research.
Research Outputs
A list of outputs from work carried out on Viper can be found on Research Outputs including publications and conference presentations. Also included is the standard acknowledgement statement we request you include in your research outputs.
If your work is missing from this list, please contact viper@hull.ac.uk with the details.
Case Studies
Endothelial Cell Biology in Health and Chronic Disease – A case study by Eamon Faulkner of The Endothelial Cell Biology in Health and Chronic Disease Research Group led by Dr. Leonid Nikitenko at the University of Hull.
Modelling Molecules In Future Oceans Using Viper – Christina Roggatz, in Dr Benoit’s Computational Chemistry Group, ran more than 500 jobs, utilising over 17,000 CPU hours between August 2016 and June 2017.
The Largest Cosmic Structures Simulated on Viper – Between Viper going live on June 28th 2016 and November 28th 2016, Dr Elke Roediger and her research group ran nearly 900 jobs on Viper, using over 715,000 CPU hours.
PhD Case Studies – A number of case studies from PhD students across the University
Research Areas
Astrophysics
Galactic Archaeology – our team within the E.A. Milne Centre for Astrophysics has pioneered the use of computational fluid dynamics married to chemical element evolution as a means to isolate and study the complex physics underpinning the formation of galaxies such as our Milky Way. Mining the fossil record of billions of stars throughout our Galaxy is one of the most audacious experiments in science today, involving an array of space and ground-based telescopes and instruments.
 |  |
Viper lies at the heart of the Milne Center’s efforts, with a 15+ year history of pushing the field in new directions. The deployment of Viper at the University of Hull heralds an exciting phase in our efforts, affording us facility access second-to-none in the world, and a resource which allows us to tackle the Grand Challenge problems which would otherwise been outside of our grasp.”
More information can be found at the E.A.Milne Centre webpages.
Biomedical Engineering
Our use of the HPC is predominantly focussed on high resolution finite element modelling of structural problems, especially of bone and similar structures.
The long term goal of the work is to develop a fully integrated multi-scale model of bone. As part of that work we are developing our own voxel-based finite element software VOX-FE, for the analysis of very large-scale, high-resolution models of bone structures. Voxel based modelling allows us to go directly from microCT scan data to finite element models with minimal loss of data and geometry simplifications thereby capturing all the complex internal and external features. It also allows inclusion of local variation in material properties based on each voxel’s grey scale value. It is these microscopic-level geometry and material property variations that explain much of the complex macroscopic behaviour of bone.
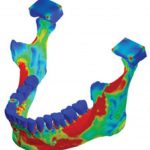 | 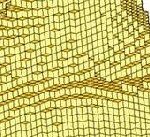 |
Commercial finite element software is typically limited to models of less than 10 million elements, using VOX-FE we have now solved models with over 200 million elements. Our target is over a billion elements. Adaptive remodelling is also under development, requiring repeated model solution, where the model adapts to the loads it experiences in the same way that bone is continually adapting in the body.
 | 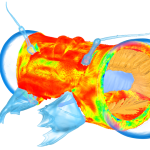 |
The number of potential applications and opportunities for VOX-FE is enormous, but so are the computer resources required, especially for the most complex, adaptive models. Viper provides those opportunities and offers the prospects of some very exciting science. For example, applications include predictive biology and virtual experimentation, and in silico design and testing of new dental and orthopaedic implants.
Chemistry
Our research in computational chemistry focuses on three main areas: molecule–surface interactions, pH effects on biological molecules, and new materials for gas adsorption. All three topics help to answer questions relating to surface adhesion, impact of ocean acidifications on marine life and carbon-capture, for example.

We develop hybrid approaches to potential energy landscape exploration and vibrational structure determination which are tailored to use both grid computing and high-performance computing (HPC). Our work on biological molecules concentrates on conformations, solvent effects, and accurate computation of nuclear magnetic resonance spectra. We also investigate molecular capture mechanisms in new materials using advanced reaction path sampling and high-end 3-D visualisation techniques.

The new HPC facility, Viper, will enable us to improve the accuracy of our models and explore larger systems while simultaneously reaching longer simulation timescales. Viper is built using the most up-to-date hardware from Intel, which will give us the opportunity to devise and test novel computational techniques that can harness the power of emerging technologies (accelerator-based HPC, for example). The capabilities of Viper will position us at the forefront of computational chemistry and help shape the future of our field of research.
Computer Sciences
Our research involves the use of GPUs, hybrids, and extreme memory configurations. Our HPC research involves running Monte Carlo tasks with 100 independent jobs, something the HPC is ideal for. Also, our research work occasionally takes us into exotic new languages, quantum computer simulations, concurrency management systems, and some low level experimentation with Intel’s latest register/SIMD capabilities.

Engineering
Our research looks with the HPC looks at large scale CFD simulations for Large Eddy Simulation of turbulent flows – applications: Combustion instability in gas turbine combustors Fires in buildings Marine vertical axis turbines Scour and erosion around offshore wind turbines.

Another engineering area are looking at the design and modelling of novel materials, process modelling and optimisation. The HPC’s involvement would look at phase field modelling and phase field crystal modelling.
EvoHull Bioinformatics
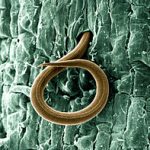
EvoHull, the Evolutionary and Environmental Genomics group, has an active research programme addressing fundamental biological questions using high throughput genomic data. They are transitioning their bioinformatics analyses to use the new HPC facilities and this will bring very substantial increase in speed and the range of activities that are possible. Genome assembly is the processing of the DNA ‘jigsaw pieces’ to create a complete picture of the organism’s genome. This is a very computationally demanding task and the university Viper HPC has included high-memory nodes for these tasks.

Root knot nematodes are one of the world’s most important crop pests, causing a 5% annual loss in world agriculture. Funded by the Natural Environment Research Council Dr Dave Lunt has established a research programme to sequence the genomes these species, providing knowledge and resources to the agricultural community. This massive genomic data set has initially been analysed using the computational resources of collaborators at Edinburgh University, but now with the investment in Viper at Hull will provide new possibilities for future research. Dr Amir Szitenberg, part of this international project, has already assembled a Root Knot Nematode genome on Viper and developed a range of novel genome analysis techniques to exploit the power of our HPC.
 |  |
The EvoHull group has developed unique biomonitoring techniques using eDNA. A key component of these revolutionary survey techniques is the rapid DNA sequence analysis made possible by Viper. The EvoHull bioinformatician Dr Christoph Hahn has written eDNA analysis software specifically to incorporate HPC analysis on Viper. Current projects are funded by Forest Research, The Environment Agency, Scottish Environmental Protection Agency, Natural Resources Wales.
Geography, Environment and Earth Sciences
We use HPC for ensemble hydrological and hydro-geomorphological models, climate modelling, climate-model downscaling and satellite image data processing, working with timescales ranging from weeks to millennia. We are also developing a new programming language for implementing this type of model on HPC (http://www.pm-lang.org/)

Languages, Linguistics & Cultures Logistics
Research in computational linguistics often relies on training a stochastic model from a dataset of examples in human natural language. The idea is to learn a mapping from a sequence of semantic symbols that represent meaning to a sequence of words that express this meaning in natural language. Such models can then be used to both understand and produce language, e.g. human-computer interaction. A common problem with state-of-the-art methods is that the features used during training are often symbolic, i.e. specific words or syntactic constructions, which heavily restricts the generalisability of a model from one domain to another.
 |  |
Recent investigations into deep learning have shown that particularly recursive neural networks, when trained from large enough datasets, lead to remarkable results in tasks such as computer vision, signal processing or machine translation. The reason is that these algorithms learn abstract feature representations that capture general patterns in the data rather than specific combinations of symbols. There is a lot of promise for the use of deep learning in computational linguistics as well, as evidenced by an increased interest from leading academic institutions and global companies, such as Google, Apple or Facebook.
A challenge to the wider uptake of these methods in academia is currently the computational resource involved in training a deep learner, which involves both non-linear computations and large volumes of training data, which can lead to weeks of training time for a single model. Using the university’s HPC Viper, we hope to learn better stochastic models of natural language by exploiting abstract commonalities from large and general-purpose natural language datasets.
Physics and Mathematics
Our research in semiconductor devices employs a sophisticated in-house developed Ensemble Monte-Carlo simulation to monitor the motion and interactions of millions of electrons. We focus predominantly on Gallium-Nitride devices – this material is currently revolutionizing the solid state lighting market and could do the same for the electronics market. A number of challenges remain, notably around heating and premature device breakdown.
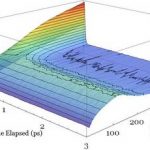 | 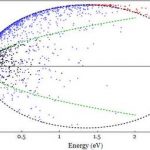 |
The new HPC facility, Viper will enable us to include additional physics to finesse our simulation tool allowing us to provide results which are of greater relevance to experimentalists and engineers. Until now we have been constrained in our science by the limitations of the hardware.

